Contents
- part 1: working with small distributions of neurons (exhaustively)
- part 2: working with larger distributions of neurons (MCMC)
- part 3: working with RP (random projection) models
- part 4: specifying a custom list of correlations
- part 5: constructing composite models
- part 6: constructing and sampling from high order Markov chains
% maxent_example.m % % Example code for the maximum entropy toolkit % Ori Maoz, July 2016: orimaoz@gmail.com, % % Note: all the "maxent." prefixes before the function calls can be omitted by commenting out the following line: %import maxent.*
part 1: working with small distributions of neurons (exhaustively)
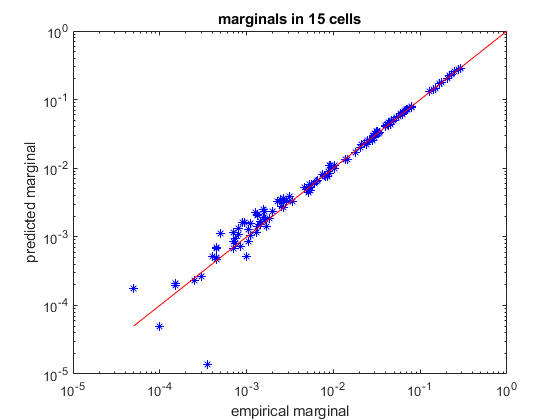
% load spiking data of 15 neurons load example15 % randomly divide it into a training set and a test set (so we can verify how well we trained) [ncells,nsamples] = size(spikes15); idx_train = randperm(nsamples,ceil(nsamples/2)); idx_test = setdiff(1:nsamples,idx_train); samples_train = spikes15(:,idx_train); samples_test = spikes15(:,idx_test); % create a k-pairwise model (pairwise maxent with synchrony constraints) model = maxent.createModel(ncells,'kising'); % train the model to a threshold of one standard deviation from the error of computing the marginals. % because the distribution is relatively small (15 dimensions) we can explicitly represent all 2^15 states % in memory and train the model in an exhaustive fashion. model = maxent.trainModel(model,samples_train,'threshold',1); % now check the kullback-leibler divergence between the model predictions and the pattern counts in the test-set empirical_distribution = maxent.getEmpiricalModel(samples_test); model_logprobs = maxent.getLogProbability(model,empirical_distribution.words); test_dkl = maxent.dkl(empirical_distribution.logprobs,model_logprobs); fprintf('Kullback-Leibler divergence from test set: %f\n',test_dkl); model_entropy = maxent.getEntropy(model); fprintf('Model entropy: %.03f empirical dataset entropy: %.03f\n', model_entropy, empirical_distribution.entropy); % get the marginals (firing rates and correlations) of the test data and see how they compare to the model predictions marginals_data = maxent.getEmpiricalMarginals(samples_test,model); marginals_model = maxent.getMarginals(model); % plot them on a log scale figure loglog(marginals_data,marginals_model,'b*'); hold on; minval = min([marginals_data(marginals_data>0)]); plot([minval 1],[minval 1],'-r'); % identity line xlabel('empirical marginal'); ylabel('predicted marginal'); title(sprintf('marginals in %d cells',ncells));
Training to threshold: 1.000 standard deviations Maximum MSE: 1.000 converged (marginals match) Standard deviations from marginals: 0.097 (mean), 0.987 (max) [9] DKL: 0.102 Kullback-Leibler divergence from test set: 0.098116 Model entropy: 6.464 empirical dataset entropy: 6.384
part 2: working with larger distributions of neurons (MCMC)
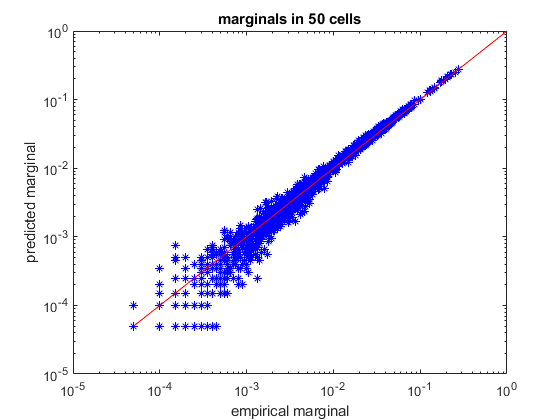
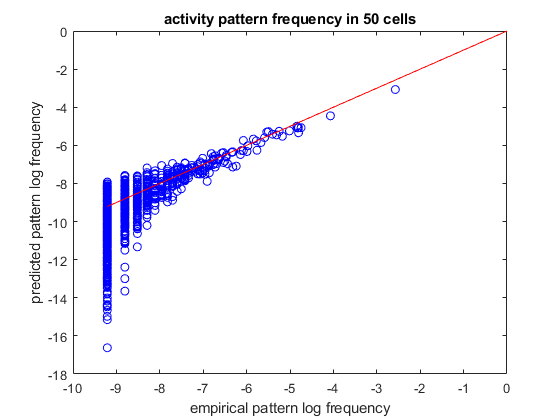
load example50 % randomly divide into train/test sets [ncells,nsamples] = size(spikes50); idx_train = randperm(nsamples,ceil(nsamples/2)); idx_test = setdiff(1:nsamples,idx_train); samples_train = spikes50(:,idx_train); samples_test = spikes50(:,idx_test); % create a pairwise maximum entropy model model = maxent.createModel(50,'pairwise'); % train the model to a threshold of 1.5 standard deviations from the error of computing the marginals. % because the distribution is larger (50 dimensions) we cannot explicitly iterate over all 5^20 states % in memory and will use markov chain monte carlo (MCMC) methods to obtain an approximation model = maxent.trainModel(model,samples_train,'threshold',1.5); % get the marginals (firing rates and correlations) of the test data and see how they compare to the model predictions. % here the model marginals could not be computed exactly so they will be estimated using monte-carlo. We specify the % number of samples we use so that their estimation will have the same amoutn noise as the empirical marginal values marginals_data = maxent.getEmpiricalMarginals(samples_test,model); marginals_model = maxent.getMarginals(model,'nsamples',size(samples_test,2)); % plot them on a log scale figure loglog(marginals_data,marginals_model,'b*'); hold on; minval = min([marginals_data(marginals_data>0)]); plot([minval 1],[minval 1],'-r'); % identity line xlabel('empirical marginal'); ylabel('predicted marginal'); title(sprintf('marginals in %d cells',ncells)); % the model that the MCMC solver returns is not normalized. If we want to compare the predicted and actual probabilities % of individual firing patterns, we will need to first normalize the model. We will use the wang-landau algorithm for % this. We chose parameters which are less strict than the default settings so that we will have a faster runtime. disp('Normalizing model...'); model = maxent.wangLandau(model,'binsize',0.1,'depth',15); % the normalization factor was added to the model structure. Now that we have a normalized model, we'll use it to % predict the frequency of activity patterns. We will start by observing all the patterns that repeated at least twice % (because a pattern that repeated at least once may grossly overrepresent its probability and is not meaningful in this % sort of analysis) limited_empirical_distribution = maxent.getEmpiricalModel(samples_test,'min_count',2); % get the model predictions for these patterns model_logprobs = maxent.getLogProbability(model,limited_empirical_distribution.words); % nplot on a log scale figure plot(limited_empirical_distribution.logprobs,model_logprobs,'bo'); hold on; minval = min(limited_empirical_distribution.logprobs); plot([minval 0],[minval 0],'-r'); % identity line xlabel('empirical pattern log frequency'); ylabel('predicted pattern log frequency'); title(sprintf('activity pattern frequency in %d cells',ncells)); % Wang-landau also approximated the model entropy, let's compare it to the entropy of the empirical dataset. % for this we want to look at the entire set, not just the set limited repeating patterns empirical_distribution = maxent.getEmpiricalModel(samples_test); % it will not be surprising to see that the empirical entropy is much lower than the model, this is because the % distribution is very undersampled fprintf('Model entropy: %.03f bits, empirical entropy (test set): %.03f bits\n',model.entropy,empirical_distribution.entropy); % generate samples from the distribution and compute their entropy. This should give a result which is must closer to % the entropy of the empirical distribution... samples_simulated = maxent.generateSamples(model,numel(idx_test)); simulated_empirical_distribution = maxent.getEmpiricalModel(samples_simulated); fprintf('Entropy of simulated data: %.03f bits\n',simulated_empirical_distribution.entropy);
Training to threshold: 1.500 standard deviations Maximum samples: 17778 maximum MSE: 3.375 361/Inf samples=5466 MSE=6.504 (mean), 57.338 (max) [43] 436/Inf samples=11581 MSE=3.315 (mean), 66.423 (max) [2] 479/Inf samples=17778 MSE=2.391 (mean), 28.954 (max) [2] 514/Inf samples=17778 MSE=1.754 (mean), 22.708 (max) [40] 549/Inf samples=17778 MSE=1.699 (mean), 28.873 (max) [12] 584/Inf samples=17778 MSE=1.642 (mean), 11.995 (max) [45] 618/Inf samples=17778 MSE=1.716 (mean), 11.184 (max) [31] 652/Inf samples=17778 MSE=1.644 (mean), 10.448 (max) [1166] 686/Inf samples=17778 MSE=1.595 (mean), 8.928 (max) [1140] 721/Inf samples=17778 MSE=1.820 (mean), 7.465 (max) [40] 756/Inf samples=17778 MSE=1.508 (mean), 7.806 (max) [12] 791/Inf samples=17778 MSE=1.491 (mean), 9.723 (max) [12] 824/Inf samples=17778 MSE=1.521 (mean), 6.340 (max) [12] 857/Inf samples=17778 MSE=1.412 (mean), 4.215 (max) [675] 891/Inf samples=17778 MSE=1.541 (mean), 6.946 (max) [1264] 925/Inf samples=17778 MSE=1.511 (mean), 5.560 (max) [142] 960/Inf samples=17778 MSE=1.695 (mean), 7.271 (max) [1151] 996/Inf samples=17778 MSE=1.601 (mean), 5.649 (max) [1151] 1031/Inf samples=17778 MSE=1.464 (mean), 4.822 (max) [1151] 1066/Inf samples=17778 MSE=1.291 (mean), 6.976 (max) [410] 1102/Inf samples=17778 MSE=1.331 (mean), 3.873 (max) [23] 1138/Inf samples=17778 MSE=1.297 (mean), 3.436 (max) [258] converged (marginals match) Normalizing model... [...............] Model entropy: 17.527 bits, empirical entropy (test set): 11.978 bits Entropy of simulated data: 12.490 bits
part 3: working with RP (random projection) models
% load spiking data of 15 neurons load example15 % randomly divide it into a training set and a test set (so we can verify how well we trained) [ncells,nsamples] = size(spikes15); idx_train = randperm(nsamples,ceil(nsamples/2)); idx_test = setdiff(1:nsamples,idx_train); samples_train = spikes15(:,idx_train); samples_test = spikes15(:,idx_test); % create a random projection model with default settings model = maxent.createModel(ncells,'rp'); % train the model to a threshold of one standard deviation from the error of computing the marginals. % because the distribution is relatively small (15 dimensions) we can explicitly represent all 2^15 states % in memory and train the model in an exhaustive fashion. model = maxent.trainModel(model,samples_train,'threshold',1); % now check the kullback-leibler divergence between the model predictions and the pattern counts in the test-set empirical_distribution = maxent.getEmpiricalModel(samples_test); model_logprobs = maxent.getLogProbability(model,empirical_distribution.words); test_dkl = maxent.dkl(empirical_distribution.logprobs,model_logprobs); fprintf('Kullback-Leibler divergence from test set: %f\n',test_dkl); % create a random projection model with a specified number of projections and specified average in-degree model = maxent.createModel(ncells,'rp','nprojections',500,'indegree',4); % train the model model = maxent.trainModel(model,samples_train,'threshold',1); % now check the kullback-leibler divergence between the model predictions and the pattern counts in the test-set empirical_distribution = maxent.getEmpiricalModel(samples_test); model_logprobs = maxent.getLogProbability(model,empirical_distribution.words); test_dkl = maxent.dkl(empirical_distribution.logprobs,model_logprobs); fprintf('Kullback-Leibler divergence from test set: %f\n',test_dkl);
Training to threshold: 1.000 standard deviations Maximum MSE: 1.000 64/Inf MSE=0.088 (mean), 4.981 (max) [65] DKL: 0.122 134/Inf MSE=0.014 (mean), 2.213 (max) [65] DKL: 0.112 converged (marginals match) Standard deviations from marginals: 0.072 (mean), 0.992 (max) [65] DKL: 0.110 Kullback-Leibler divergence from test set: 0.125975 Training to threshold: 1.000 standard deviations Maximum MSE: 1.000 09/Inf MSE=56.463 (mean), 7086.771 (max) [69] DKL: 0.243 29/Inf MSE=0.644 (mean), 58.533 (max) [69] DKL: 0.121 49/Inf MSE=0.126 (mean), 15.019 (max) [364] DKL: 0.108 69/Inf MSE=0.075 (mean), 8.094 (max) [364] DKL: 0.103 89/Inf MSE=0.031 (mean), 3.065 (max) [364] DKL: 0.100 109/Inf MSE=0.022 (mean), 3.862 (max) [125] DKL: 0.097 129/Inf MSE=0.013 (mean), 2.741 (max) [125] DKL: 0.095 149/Inf MSE=0.008 (mean), 1.318 (max) [125] DKL: 0.094 converged (marginals match) Standard deviations from marginals: 0.083 (mean), 0.997 (max) [125] DKL: 0.093 Kullback-Leibler divergence from test set: 0.108484
part 4: specifying a custom list of correlations
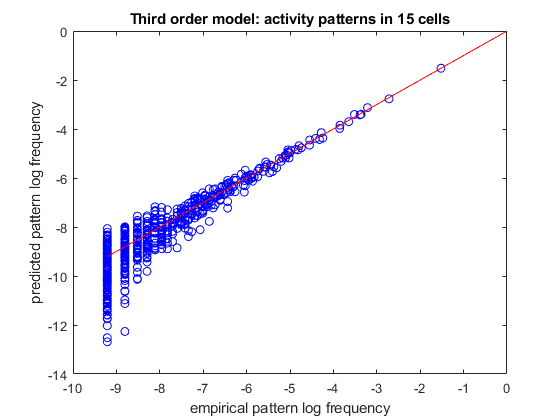
% load spiking data of 15 neurons load example15 % randomly divide it into a training set and a test set (so we can verify how well we trained) [ncells,nsamples] = size(spikes15); idx_train = randperm(nsamples,ceil(nsamples/2)); idx_test = setdiff(1:nsamples,idx_train); samples_train = spikes15(:,idx_train); samples_test = spikes15(:,idx_test); % create a model with first, second, and third-order correlations. (third-order model) % we will do this by specifying a list of all the possible combinations of single factors, pairs and triplets correlations = cat(1,num2cell(nchoosek(1:ncells,1),2), ... num2cell(nchoosek(1:ncells,2),2),... num2cell(nchoosek(1:ncells,3),2)); model = maxent.createModel(ncells,'highorder',correlations); % train it model = maxent.trainModel(model,samples_train,'threshold',1); % use the model to predict the frequency of activity patterns. % We will start by observing all the patterns that repeated at least twice (because a pattern that repeated at least % once may grossly overrepresent its probability and is not meaningful in this sort of analysis) limited_empirical_distribution = maxent.getEmpiricalModel(samples_test,'min_count',2); % get the model predictions for these patterns model_logprobs = maxent.getLogProbability(model,limited_empirical_distribution.words); % nplot on a log scale figure plot(limited_empirical_distribution.logprobs,model_logprobs,'bo'); hold on; minval = min(limited_empirical_distribution.logprobs); plot([minval 0],[minval 0],'-r'); % identity line xlabel('empirical pattern log frequency'); ylabel('predicted pattern log frequency'); title(sprintf('Third order model: activity patterns in %d cells',ncells));
Training to threshold: 1.000 standard deviations Maximum MSE: 1.000 26/Inf MSE=3.765 (mean), 651.384 (max) [12] DKL: 0.280 55/Inf MSE=1.461 (mean), 42.061 (max) [8] DKL: 0.145 84/Inf MSE=0.555 (mean), 25.381 (max) [13] DKL: 0.110 113/Inf MSE=0.251 (mean), 20.876 (max) [2] DKL: 0.098 142/Inf MSE=0.125 (mean), 5.159 (max) [2] DKL: 0.092 171/Inf MSE=0.089 (mean), 5.211 (max) [278] DKL: 0.089 199/Inf MSE=0.047 (mean), 1.658 (max) [501] DKL: 0.087 228/Inf MSE=0.028 (mean), 1.828 (max) [12] DKL: 0.085 255/Inf MSE=0.025 (mean), 1.676 (max) [501] DKL: 0.084 282/Inf MSE=0.023 (mean), 1.690 (max) [501] DKL: 0.083 converged (marginals match) Standard deviations from marginals: 0.116 (mean), 0.990 (max) [501] DKL: 0.083
part 5: constructing composite models
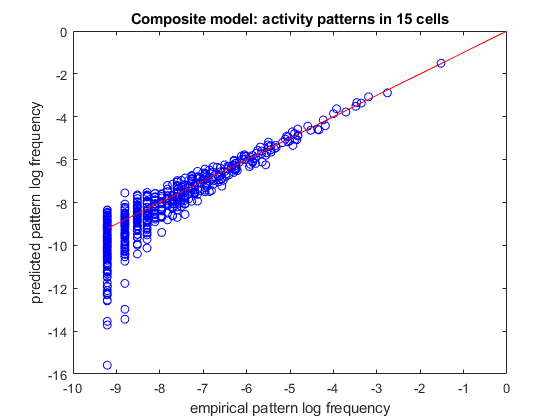
% load spiking data of 15 neurons load example15 % randomly divide it into a training set and a test set (so we can verify how well we trained) [ncells,nsamples] = size(spikes15); idx_train = randperm(nsamples,ceil(nsamples/2)); idx_test = setdiff(1:nsamples,idx_train); samples_train = spikes15(:,idx_train); samples_test = spikes15(:,idx_test); % create a model with independent factors, k-synchrony and third-order correlations % we will do this by initializing 3 separate models and then combining them to a single model third_order_correlations = num2cell(nchoosek(1:ncells,3),2); model_indep = maxent.createModel(ncells,'indep'); model_ksync = maxent.createModel(ncells,'ksync'); model_thirdorder = maxent.createModel(ncells,'highorder',third_order_correlations); model = maxent.createModel(ncells,'composite',{model_indep,model_ksync,model_thirdorder}); % train it model = maxent.trainModel(model,samples_train,'threshold',1); % use the model to predict the frequency of activity patterns. % We will start by observing all the patterns that repeated at least twice (because a pattern that repeated at least % once may grossly overrepresent its probability and is not meaningful in this sort of analysis) limited_empirical_distribution = maxent.getEmpiricalModel(samples_test,'min_count',2); % get the model predictions for these patterns model_logprobs = maxent.getLogProbability(model,limited_empirical_distribution.words); % nplot on a log scale figure plot(limited_empirical_distribution.logprobs,model_logprobs,'bo'); hold on; minval = min(limited_empirical_distribution.logprobs); plot([minval 0],[minval 0],'-r'); % identity line xlabel('empirical pattern log frequency'); ylabel('predicted pattern log frequency'); title(sprintf('Composite model: activity patterns in %d cells',ncells));
Training to threshold: 1.000 standard deviations Maximum MSE: 1.000 30/Inf MSE=0.484 (mean), 26.973 (max) [12] DKL: 0.122 62/Inf MSE=0.152 (mean), 4.076 (max) [24] DKL: 0.111 94/Inf MSE=0.062 (mean), 2.372 (max) [150] DKL: 0.106 126/Inf MSE=0.030 (mean), 1.534 (max) [458] DKL: 0.104 159/Inf MSE=0.025 (mean), 4.956 (max) [24] DKL: 0.103 192/Inf MSE=0.015 (mean), 1.924 (max) [24] DKL: 0.101 225/Inf MSE=0.011 (mean), 1.095 (max) [458] DKL: 0.100 converged (marginals match) Standard deviations from marginals: 0.097 (mean), 1.000 (max) [458] DKL: 0.100
part 6: constructing and sampling from high order Markov chains
train a time-dependent model
% load spiking data of 15 neurons load example15_spatiotemporal ncells = size(spikes15_time_dependent,1); history_length = 2; % Create joint words of (x_t-2,x_t-1,x_t). xt = []; for i = 1:(history_length+1) xt = [xt;spikes15_time_dependent(:,i:(end-history_length-1+i))]; end % create a spatiotemporal model that works on series of binary words: (x_t-2,x_t-1,x_t). % this essentially describes the probability distribution as a second-order Markov process. % We will model the distribution with a composite model that uses firing rates, total synchrony in the last 3 time bins % and pairwise correlations within the current time bin and pairwise correlations between the current activity of a cell % and the previous time bin. time_dependent_ncells = ncells*(history_length+1); inner_model_indep = maxent.createModel(time_dependent_ncells,'indep'); % firing rates inner_model_ksync = maxent.createModel(time_dependent_ncells,'ksync'); % total synchrony in the population % add pairwise correlations only within the current time bin second_order_correlations = num2cell(nchoosek(1:ncells,2),2); temporal_matrix = reshape(1:time_dependent_ncells,[ncells,history_length+1]); temporal_interactions = []; % add pairwise correlations from the current time bin to the previous time step for i = 1:(history_length) temporal_interactions = [temporal_interactions;temporal_matrix(:,[i,i+1])]; end % bunch of all this together into one probabilistic model temporal_interactions = num2cell(temporal_interactions,2); inner_model_pairwise = maxent.createModel(time_dependent_ncells,'highorder',[second_order_correlations;temporal_interactions]); mspatiotemporal = maxent.createModel(time_dependent_ncells,'composite',{inner_model_indep,inner_model_ksync,inner_model_pairwise}); % train the model on the concatenated words disp('training spatio-temporal model...'); mspatiotemporal = maxent.trainModel(mspatiotemporal,xt); % sample from the model by generating each sample according to the history. % for this we need to generate the 3n-dimensional samples one by one, each time fixing two-thirds of the code word % corresponding to time t-2 and t-1 and sampling only from time t. disp('sampling from spatio-temporal model...'); x0 = uint32(xt(:,1)); xspatiotemporal = []; nsamples = 10000; for i = 1:nsamples % get next sample. we will use burn-in of 100 each step to ensure that we don't introduce time-dependent stuff % related to the sampling process itself. xnext = maxent.generateSamples(mspatiotemporal,1,'fix_indices',1:(ncells*history_length),'burnin',100,'x0',x0); generated_sample = xnext(((ncells*history_length)+1):end,1); % shift the "current" state by one time step x0 = [x0((ncells+1):end,:);generated_sample]; % add the current output xspatiotemporal = [xspatiotemporal,generated_sample]; end % plot the result display_begin = 2000; nsamples_to_display = 300; % plot the actual raster pos = [400,600,700,250]; figure('Position',pos); subplot(2,1,1); pos = get(gca, 'Position');pos(1) = 0.055;pos(3) = 0.9;set(gca, 'Position', pos); imshow(~spikes15_time_dependent(:,display_begin+(1:nsamples_to_display))); title('Actual data'); % plot raster sampled from a spatiotemporal model subplot(2,1,2); pos = get(gca, 'Position');pos(1) = 0.055;pos(3) = 0.9;set(gca, 'Position', pos); imshow(~xspatiotemporal(:,display_begin+(1:nsamples_to_display))); title('Synthetic data (2nd-order Markov)');
training spatio-temporal model... Training to threshold: 1.300 standard deviations Maximum samples: 47335 maximum MSE: 2.535 283/Inf samples=2488 MSE=19.036 (mean), 47.786 (max) [48] 357/Inf samples=5250 MSE=9.391 (mean), 33.829 (max) [103] 401/Inf samples=8163 MSE=6.096 (mean), 21.037 (max) [212] 431/Inf samples=11016 MSE=4.558 (mean), 17.024 (max) [222] 455/Inf samples=14000 MSE=3.666 (mean), 10.921 (max) [52] 474/Inf samples=16923 MSE=3.058 (mean), 7.152 (max) [59] 490/Inf samples=19851 MSE=2.590 (mean), 7.018 (max) [99] 504/Inf samples=22824 MSE=2.239 (mean), 8.185 (max) [101] 516/Inf samples=25725 MSE=1.969 (mean), 9.438 (max) [101] 527/Inf samples=28707 MSE=1.797 (mean), 8.821 (max) [101] 537/Inf samples=31713 MSE=1.656 (mean), 7.257 (max) [101] 546/Inf samples=34690 MSE=1.598 (mean), 8.343 (max) [212] 554/Inf samples=37567 MSE=1.502 (mean), 8.692 (max) [212] 562/Inf samples=40683 MSE=1.403 (mean), 9.427 (max) [212] 570/Inf samples=44058 MSE=1.310 (mean), 9.120 (max) [212] 577/Inf samples=47239 MSE=1.227 (mean), 8.797 (max) [212] 584/Inf samples=47335 MSE=1.150 (mean), 8.708 (max) [212] 591/Inf samples=47335 MSE=1.082 (mean), 8.041 (max) [212] 598/Inf samples=47335 MSE=1.008 (mean), 7.813 (max) [212] 605/Inf samples=47335 MSE=0.978 (mean), 6.697 (max) [212] 612/Inf samples=47335 MSE=0.925 (mean), 5.669 (max) [212] 619/Inf samples=47335 MSE=0.902 (mean), 5.225 (max) [212] 626/Inf samples=47335 MSE=0.905 (mean), 4.294 (max) [212] 633/Inf samples=47335 MSE=0.908 (mean), 3.184 (max) [212] converged (marginals match) sampling from spatio-temporal model...
